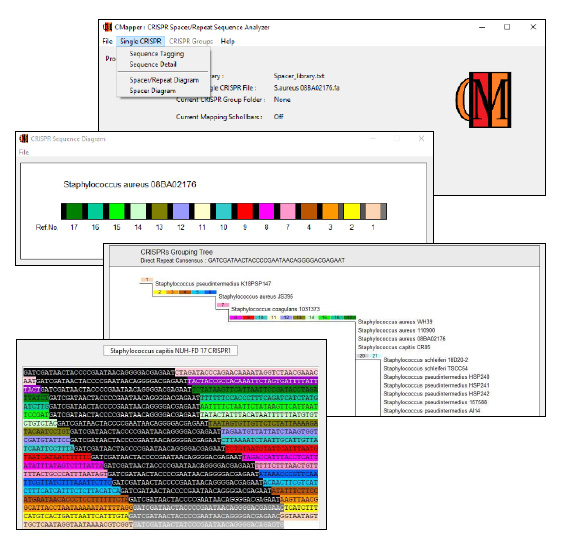
CMapper : CRISPR Spacer/Repeat Sequence Analyzer (version 2.0)
Copyright (c) 2023
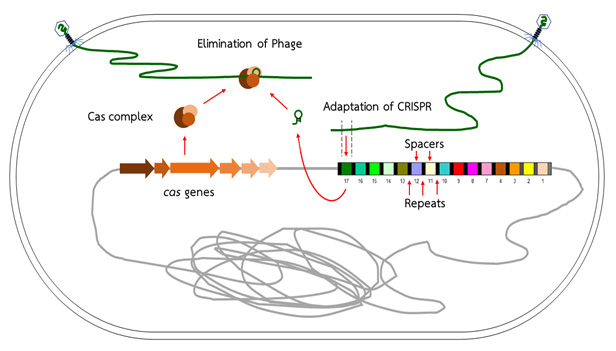
CMapper : CRISPR Spacer/Repeat Sequence Analyzer is a bioinformatics program that helps analyze the structure of CRISPR spacer/repeat region. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a microorganism system with a function similar to that of adaptive immunity found in high-level animals. The CRISPR system involves cas genes (CRISPR associated genes) and a CRISPR region. The genes in the cas system express enzymes that facilitate the defense and the adaptation of the CRISPR region against subsequent threats from the same enemy. The CRISPR region consists of 2 types of DNA sequences called direct repeat and spacer. Direct repeats are a series of identical base sequences that appear repeatedly in the same direction in CRISPR region. Each of the two repeat sequences is interspaced with a spacer sequence, which acquired from virus, plasmid, or foreign DNA attackers. The new retrieved spacer is then added next to the last added spacer at the end of CRISPR region. The base sequence of each spacer differs from other spacers in the same CRISPR system, indicates the history of various types of enemies previously confronted. Organization of CRISPR sequences form the whole group of microorganisms may shed light on their relations, sources, and history.
CRISPR-Cas system
Adapted from : Marraffini LA (2015). CRISPR-Cas immunity in prokaryotes. Nature; 526: 55-61.
Installation
CMapper : CRISPR Spacer/Repeat Sequence Analyzer has been developed as a standalone program and is available for download on three platforms: Windows, Mac OSX, and Linux. You can obtain the program using the following links:
For Windows users:CMapper_v2.0_win.rar – Google Drive
For Mac OSX users: CMapper_v2.0_mac.zip – Google Drive
For Linux users: CMapper_v2.0_linux.zip – Google Drive
After downloading and unzipping the program, Windows and Mac OSX users can simply click on the “CMapper" file to run the application. On the Linux platform, if double-clicking doesn’t work, you can run the program directly from the terminal. Open the terminal, type “. /CMapper" (without quotes), and press Enter/Return to execute the command. The CMapper menu will then appear as a graphical interface.
When running CMapper on any operating system, you may encounter security notifications related to computer viruses or require permission. In such cases, you will need to unlock the security or grant permission for the application file to run on your system.
The CMapper package also includes samples of a spacer library and a folder containing three CRISPR sequences. These sequences represent bacterial strains obtained from the NCBI website. It’s important to note that these samples are provided solely for demonstration purposes, serving as examples to showcase the functionality and capabilities of the CMapper program.
Please keep in mind that CMapper is still in development. We are continuously working on enhancing the program by implementing improved algorithms, which will be incorporated in future versions. If you encounter any issues or have suggestions for improvement, we encourage you to report them via the provided email address. We value and welcome all feedback and comments.
Plykaeow Chaibenjawong
Department of Microbiology and Parasitology
Faculty of Medical Science
Naresuan University, Thailand
Contact email: plykaeow@hotmail.com, plykaeowc@nu.ac.th

Copyright (c) 2023
All Rights Reserved.
(c) CMapper : CRISPR Spacer/Repeat Sequence Analyzer 2023 – All rights reserved
![]()